0
Gene coverage
Our product catalog includes 2 assay databases totaling up to almost one million assays, both covering almost 99% of the human protein coding exome. One database,
encompassing over 285,000 assays with amplicon lengths ranging from 350 to 750 bp, is intended for high-quality DNA samples. The second database is focused on
degraded DNA samples - such as formalin fixed parafin embedded (FFPE) samples - and contains over 530,000 assays having lengths between 125 and 275 bp. Both
databases were generated using our state-of-the-art primer design pipeline capable of generating robust and high-quality assays.
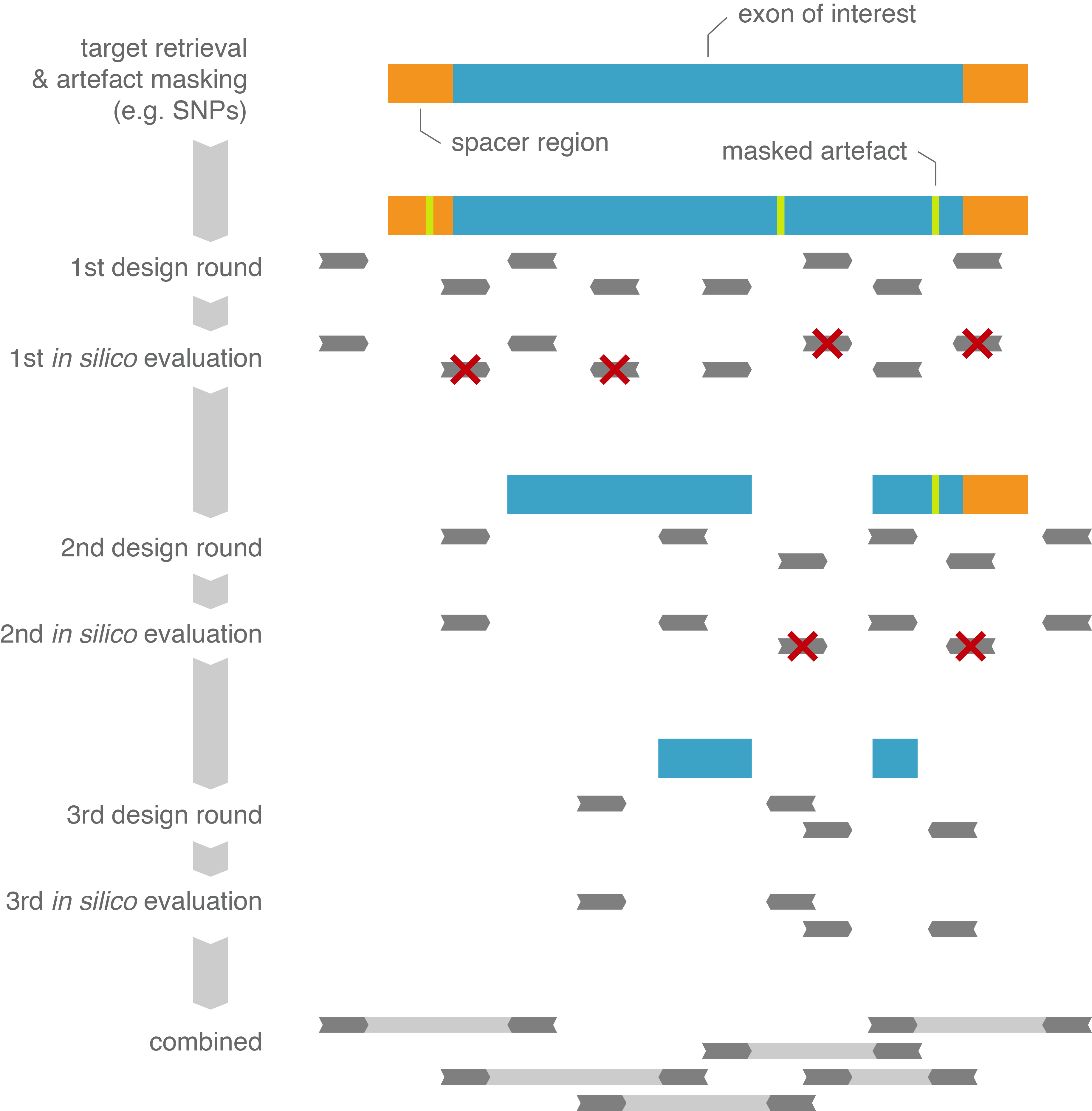
Figure 1 : Workflow of the pxlence' primer design pipeline, with the corresponding relaxation cascade and in silico evaluations
Prior to assay design, the exons of the canonical transcript for each gene were selected as target. Each target region was further extended at its 3' and 5' ends with a 25 bp spacer region to ensure
sufficient sequencing quality at the start of the exonic target region (Figure 1). This is mostly of interest for Sanger sequencing, the quality at the beginning of a read is known to be sub-optimal. Resulting
regions were then fed into our primer design pipeline.
Since our pipeline takes into account several quality parameters such as specificity and SNP/secondary structure content in primer annealing sites, generating assays using only the most stringent
design parameters would result in ill-covered targets and remaining gaps. In this context, expecially the ever increasing number of SNP scattered across the exome is making it difficult to locate
suitable primer annealing sites. To prevent this, our pipeline is equiped with an optimized relaxation cascade, which enables step-wise relaxation of design parameters
in a controled manner. This workflow, combining design parameter relaxation with stringent in silico quality evaluations, allows us to generate high-quality
assays while ensuring a maximal target coverage using a minimal number of assays (Figure 1). More details on the primer design tool and associated relaxation cascade can be found in our "High-throughput
PCR assay design for targeted resequencing using primerXL" paper (BMC Bioinformatics, 2017 ).