0
Assay specificity
In our paper "Single-nucleotide polymorphisms and other mismatches reduce performance of quantitative PCR assays"
(Clinical Chemistry, 2013 ),
we determined the impact of nucleotide mismatches in primer annealing sites on PCR efficiency. There is a negative correlation between
the number of mismatches and PCR assay performance, with higher numbers of mismatches resulting in reduced or absent amplification.
The same insights can also be be used for in silico prediction the specificity (selectivity) of a primer pair. As an integral part of our primer design engine, the potential of each
candidate assay to generate non-specific (off-target) products, is assessed by aligning the primer sequences to the genome or transcriptome while allowing
up to three mismatches per primer annealing site (and taking into account the maximum allowed product length). By collecting all possible annealing sites -
both the perfect and imperfect (non-specific) ones - and tracking the position of mismatches in them, a score depicting the amplification potential of
each site can be calculated. This strategy enables the selection of the best assays - i.e. guaranteeing no or minimal off-target
amplification - and allows our customers to critically appreciate the quality of a ConfirmPCRTM assay before using it in the lab.
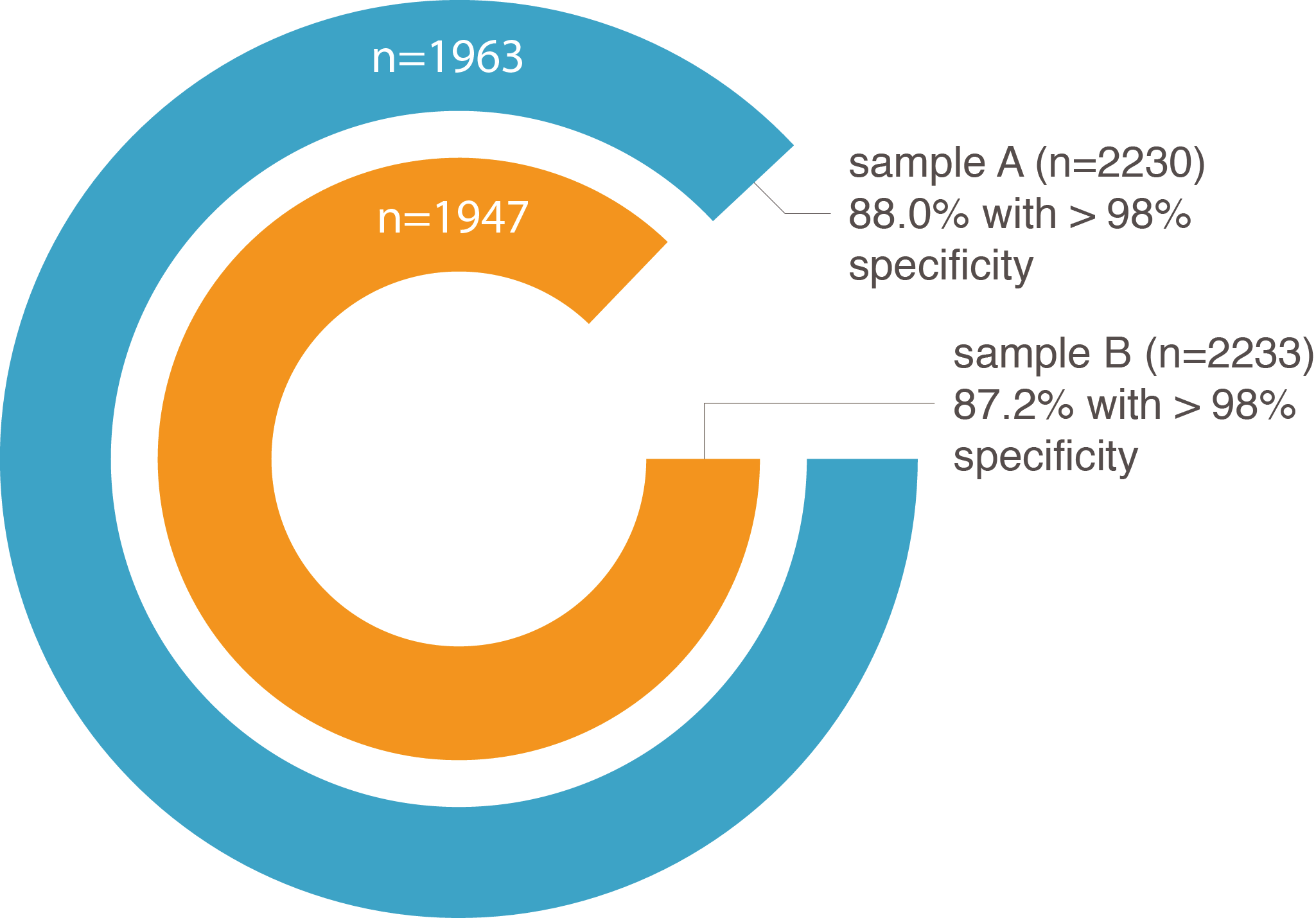
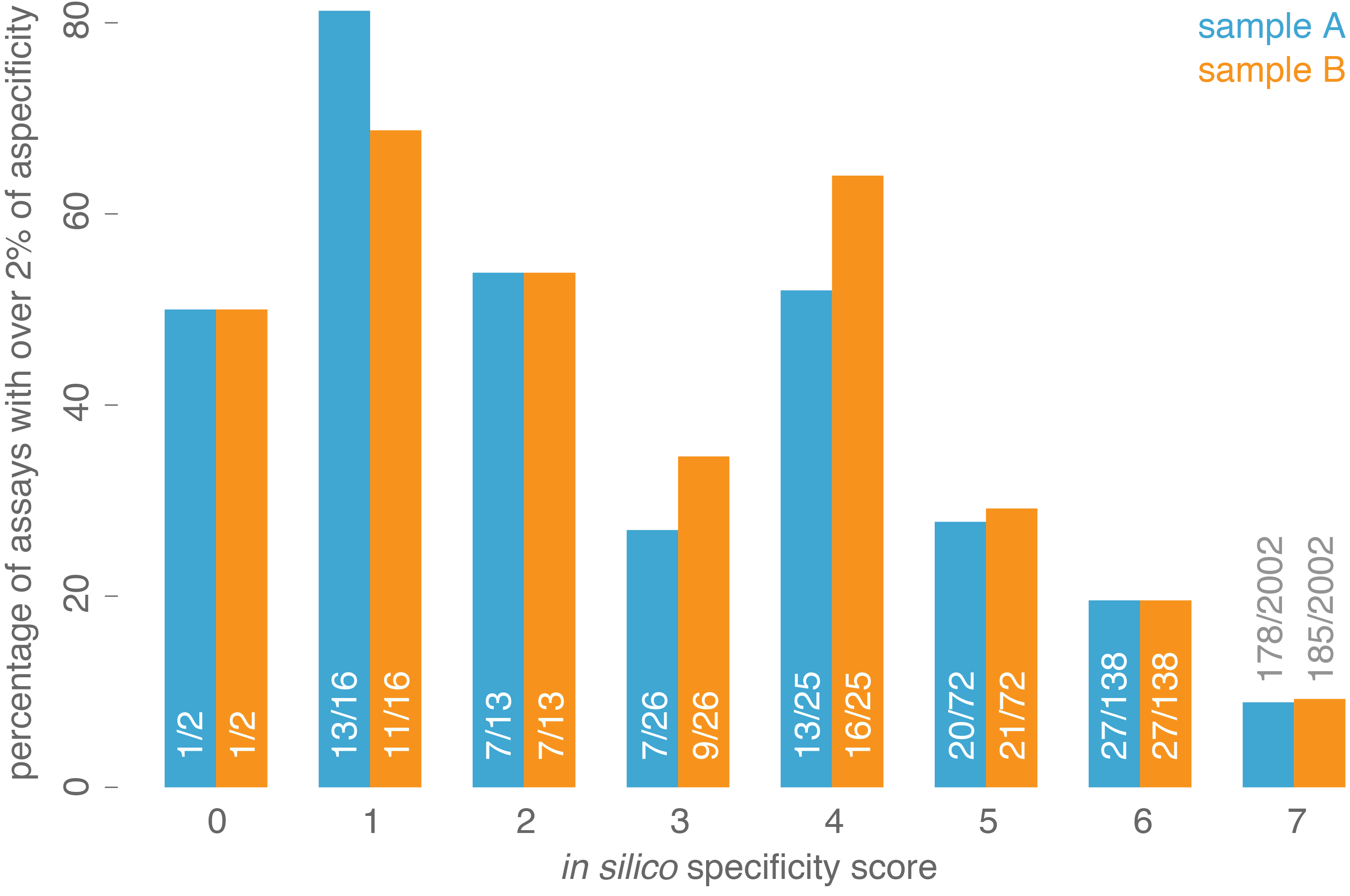
Figure 1 : Percentage of ConfirmPCRTM assays with more than 98% of the reads mapping to on-target regions
Figure 2 : Percentage of assays showing more than 2% non-specific coverage, grouped by our in silico specificity score. Numbers (x/y) in the bars
indicate the number of assays with more than 2% non-specificity (x) across all assays having the corresponding in silico
specificity score (y).
The validity of our in silico specificity score, implemented in our state-of-the-art primer design pipeline,
was empirically proven by evaluating almost 2300 assays in two DNA samples (for more details, see the paper
in the knowledge center of our website). Following standard PCR, the resulting amplicons were sequenced and their on- and off-target coverage determined. The large majority of the assays (88%) demonstrated less than 2% off-target coverage
(Figure 1). In addition, a good correlation could be shown between the calculated specificity score (ranging from 1 to 7, low to high degree of specificity) and the percentage of
assays having more than 2% aspecific coverage (Figure 2), indicating that assays with higher specificity scores tend to result in less aspecific coverage. Together,
these data illustrate once more the quality of our assays and the strength of our in silico
specificity assessment workflow and primer design pipeline.